The Onso system
SETTING A NEW STANDARD FOR SHORT READS WITH THE ONSO SYSTEM
The Onso system is an innovative benchtop short-read DNA sequencing platform with an extraordinary level of accuracy using PacBio sequencing by binding (SBB) technology. Ultra-high Q40+ data quality allows you to break through limits of detection.

Higher accuracy
Unique SBB technology delivers 15× higher accuracy for sensitive and specific characterization of low complexity regions with 90% Q40+. Fewer errors mean fewer false positives and more biological insight.
Improved sensitivity
Using fewer reads, the Onso system lowers the limit of detection, which drives the sensitivity of applications like liquid biopsy. This enables the identification of variants missed by other shortread sequencing platforms
Efficiency
Q40+ delivers high quality results at lower read depths, enabling scalable sequencing while reducing time and cost. Reduced need for unique molecular identifiers (UMIs) lowers workflow complexity and cost.

A groundbreaking new sequencer
The Onso system is a scalable and flexible benchtop platform that gives you the flexibility to integrate existing short-read tools. Powered by the innovative sequencing by binding (SBB®) technology, the Onso system delivers unprecedented accuracy and sensitivity to advance your critical and groundbreaking research.
15× higher accuracy than SBS sequencers
SBB chemistry delivers extraordinary levels of accuracy and sensitivity to confidently identify rare variants missed by other short-read sequencing platforms.
Less sequencing required
At Q30, the amount of coverage needed for certain applications (e.g. liquid biopsy research) is not economically feasible. The Onso system’s Q40+ performance provides high quality results at up to four fold less read depth than SBS.
Power through difficult to sequence regions of the genome
Near-perfect sequencing provides researchers accurate characterization of low complexity, highly repetitive regions for achieving more error-free genomes.
Google Health seamless integrations with existing laboratory processes the DeepConsensus on board
Third-party sample and library prep solutions are supported on Onso through a simple library conversion process. The Onso FASTQ file output ensures compatibility with downstream secondary and tertiary short-read analysis tools.
Sequencing by binding (SBB) for use on the Onso sequencing system
A major contributor to the Onso system’s highly accurate reads is the chemistry. Proprietary sequencing by binding (SBB) chemistry uses native nucleotides, scarless incorporation, and optimized conditions for binding and extension. These innovations result in data with decreased errors relative to competing platforms and, as a result, allow detection of rare variants below the limit of detection of existing platforms as well as resolving regions in the genome. We believe SBB will provide new opportunities for advancement of biology through highly accurate next generation sequencing.
Industry-leading sensitivity for low-frequency allele detection in liquid biopsy*
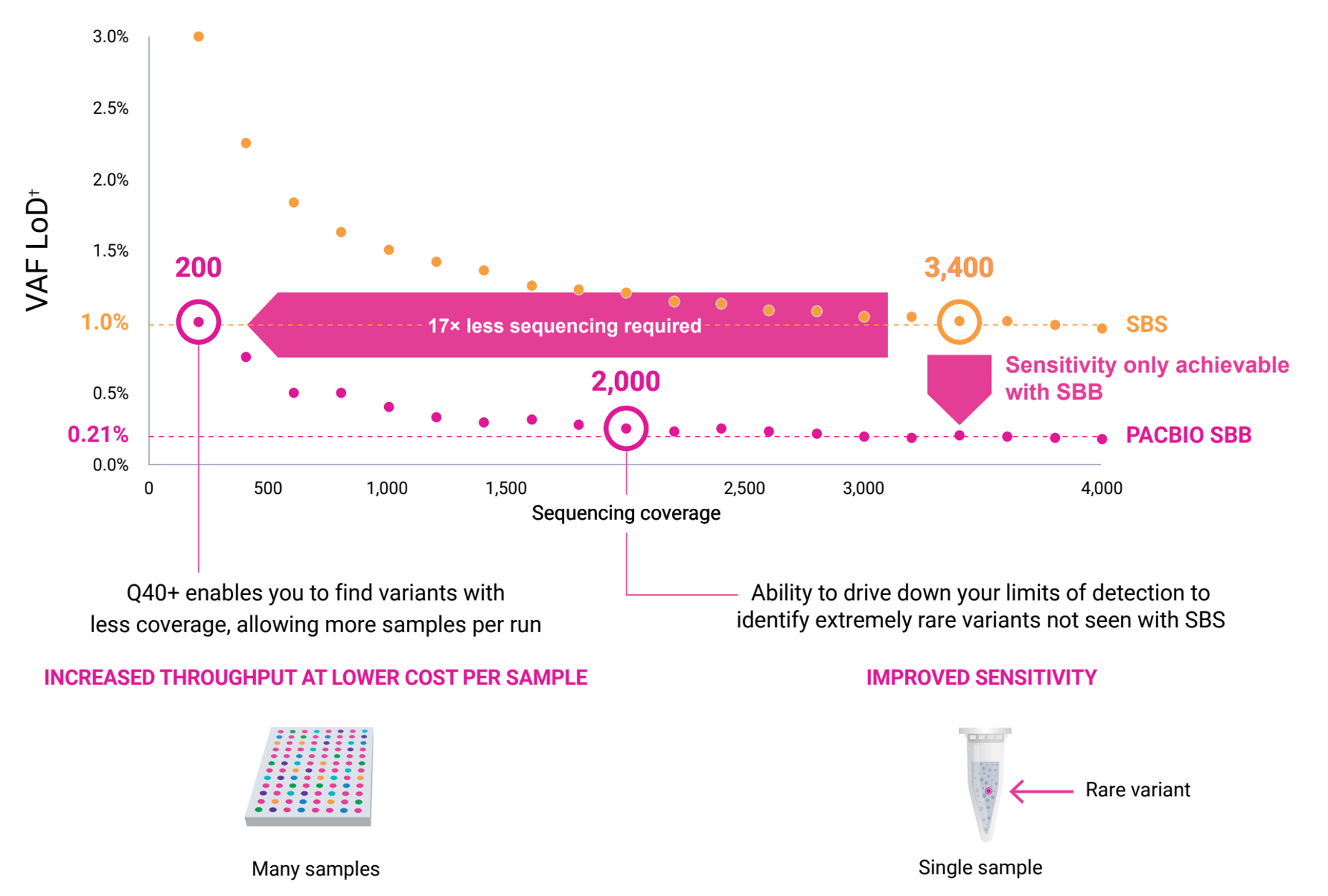
* Analytical validation (TP53 plasmid spike-in) of hybrid capture–based NGS genomic profiling of cell-free circulating tumor DNA
† Variant allele frequency (VAF) limit of detection (LoD) at 99.99% specificity for >90% of the sites
Onso system specifications
The Onso system sequencing consumables contain a flow cell (dual lane), clustering reagents, and sequencing reagent pack for supporting 200 or 300 cycles of sequencing and dual index reads.
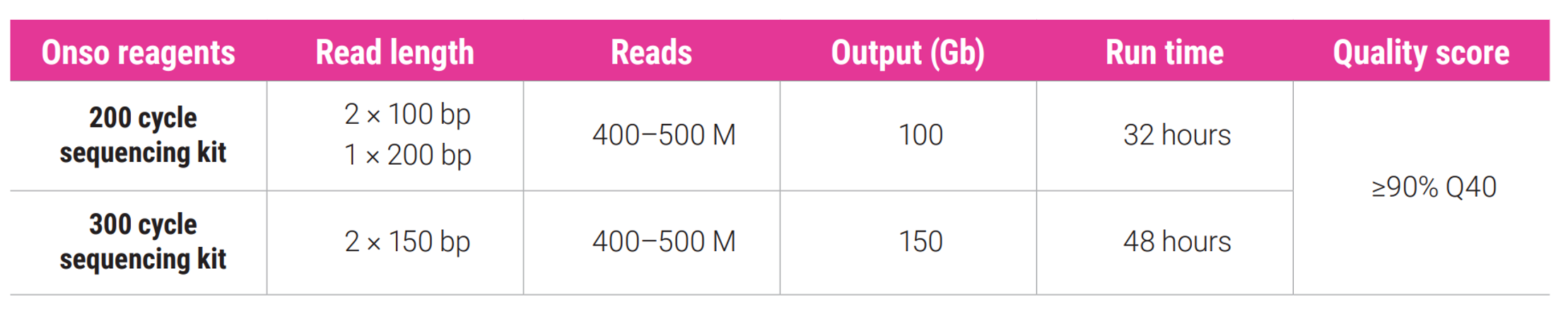
Application specifications
The Onso system is a midrange short-read sequencer capable of supporting a wide range of applications required for most laboratories.
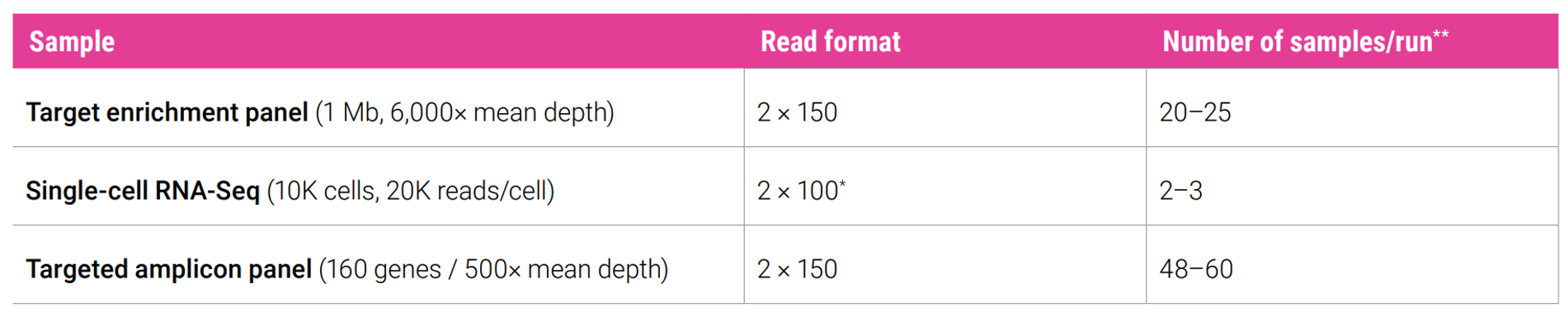
* Application-specific read format varies; supported by the Onso 200 cycle sequencing kit
** Numbers shown are estimates based on expected output per kit. Actual number of samples will vary depending on sample type, quality, and experimental objectives
What can you do with the Onso system?

Cancer research
Enable development of screening, monitoring, and therapy selection

Gene editing
Confirmation of editing outcomes and biomarker discovery

Exome/panels
Validated WES using Twist Bioscience hybrid capture protocol

Single cell
Single cell aplications supported
